pkgs <- c("ggthemes", "jhtools", "glue", "ggsci", "patchwork", "tidyverse",
"circlize", "ComplexHeatmap", "SummarizedExperiment", "jhuanglabRNAseq",
"viridis","ggrepel", "tidygraph","ggraph")
for (pkg in pkgs){
suppressPackageStartupMessages(library(pkg, character.only = T))
}
out_dir <- "./results/sup_figure12" %>% checkdir
in_dir <- "./data" %>% checkdirsup_figure12
sup_figure12A
immue_cells <- glue("{in_dir}/immue_cell.rds") %>% read_rds
group_color <- glue("{in_dir}/group_color.rds") %>% read_rds
p <- ggplot(immue_cells,
aes(x = sample_label, y = value, color = sample_label)) +
geom_boxplot(outlier.size = 0.5) +
scale_color_manual(values = group_color) +
theme_few() +
facet_wrap(~name, nrow = 2, scales = "free_y") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5)) +
theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_text(size = 7, colour = "black"),
axis.ticks = element_line(linewidth = 0.1, colour = "black"),
axis.ticks.length = unit(0.05, "lines"),
legend.box.margin = margin(0,0,0,0),
legend.box.spacing = unit(1.5, "mm"))
pdf(glue("{out_dir}/figS12A.pdf"), width = 6.7, height = 2.5)
print(p)
d <- dev.off()
p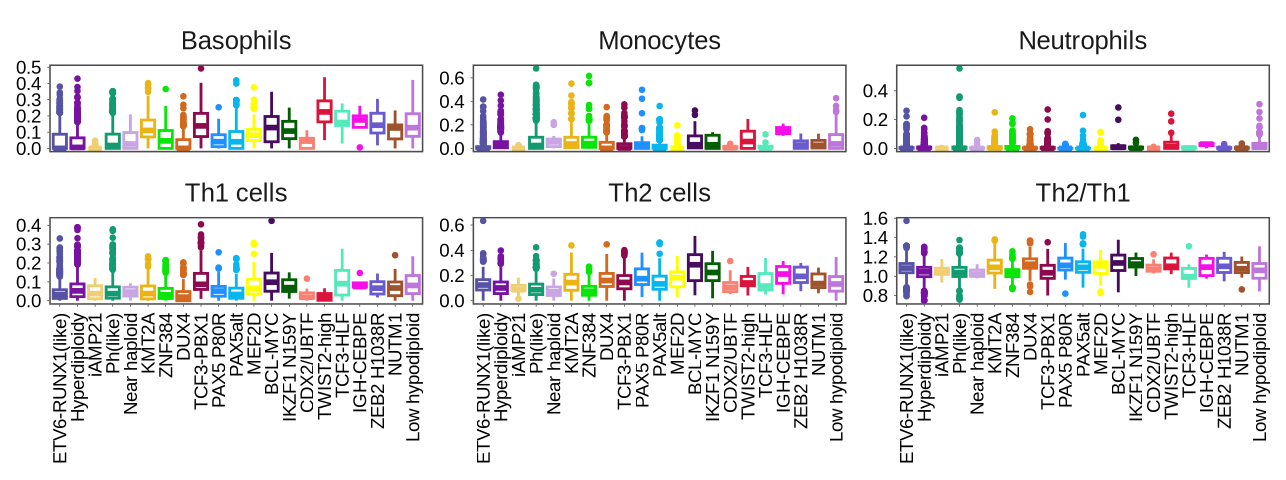
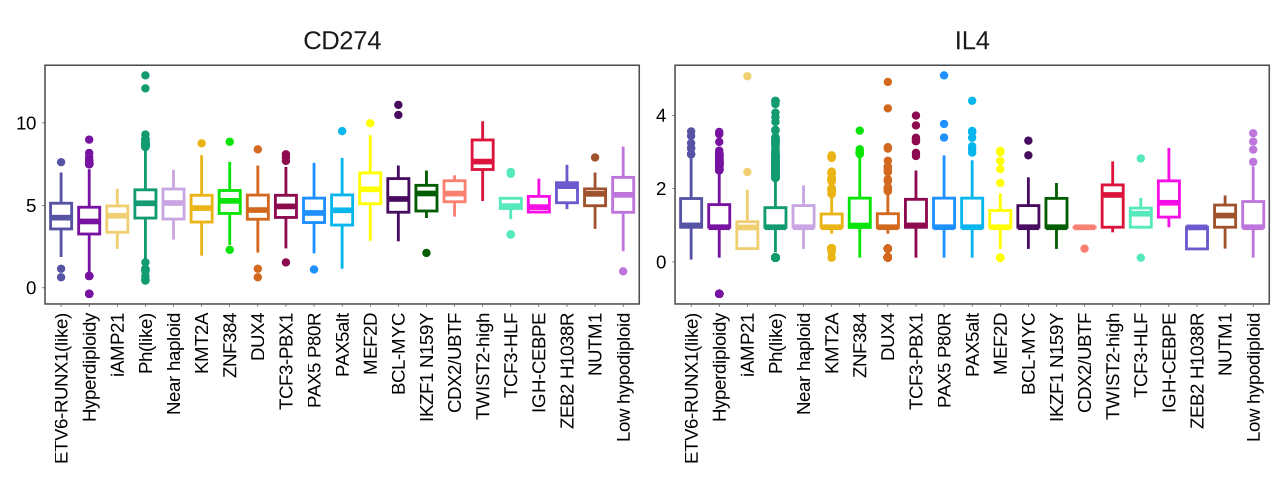
sup_figure12B
immue_gene <- glue("{in_dir}/immue_gene.rds") %>% read_rds
group_color <- glue("{in_dir}/group_color.rds") %>% read_rds
p <- ggplot(immue_gene,
aes(x = sample_label, y = value, color = sample_label)) +
geom_boxplot(outlier.size = 0.8) +
scale_color_manual(values = group_color) +
theme_few() +
facet_wrap(~name, nrow = 1, scale = "free_y") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5)) +
theme(legend.position = "none",
axis.title = element_blank(),
axis.text = element_text(size = 7, colour = "black"),
axis.ticks = element_line(linewidth = 0.1, colour = "black"),
axis.ticks.length = unit(0.05, "lines"),
legend.box.margin = margin(0,0,0,0),
legend.box.spacing = unit(1.5, "mm"))
pdf(glue("{out_dir}/figS12B.pdf"), width = 6.7, height = 2.5)
print(p)
d <- dev.off()
p