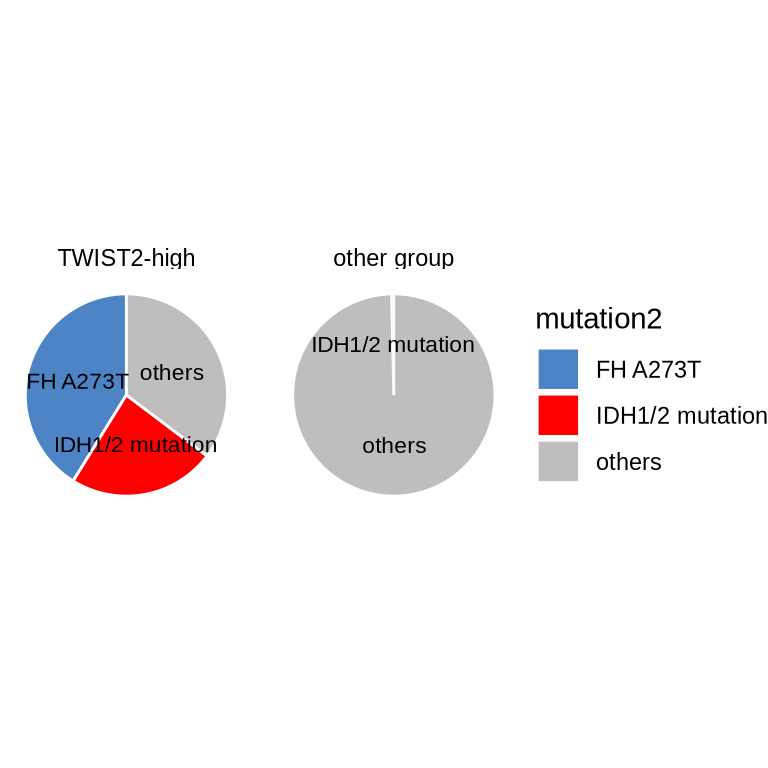
sample_frame <- read_rds(glue("{in_dir}/pie_group.rds"))
totalcod <- glue("{in_dir}/twist2_idh_colod.rds") %>% read_rds
sample_frame <- sample_frame %>%
mutate(twist2_label = case_when(
color_label == "TWIST2-high" ~ "TWIST2-high",
T ~ "other group"
),
mutation2 = case_when(
str_detect(mutation, pattern = "IDH") ~ "IDH1/2 mutation",
str_detect(mutation, pattern = "FH") ~ "FH A273T",
T ~ "others"),
twist2_label = factor(twist2_label, levels = c("TWIST2-high","other group")),
color_label = factor(color_label, levels = totalcod)) %>%
group_by(mutation2, twist2_label) %>%
summarise(n = n()) %>%
group_by(twist2_label) %>%
mutate(prop = n/sum(n))
a <- ggplot(sample_frame, aes(x = "", y = prop, fill = mutation2)) +
geom_bar(width = 1, stat = "identity", color = "white") +
geom_text(mapping = aes(x = "", y = prop, label = mutation2), position = position_stack(vjust= 0.5 ), size = 3)+
coord_polar("y", start = 0)+
scale_fill_manual(values = c("IDH1/2 mutation" = "red", "FH A273T" = "#4D84C5", "others" = "grey")) +
theme_void()+
facet_wrap(~twist2_label)
pdf(glue("{out_dir}/figure3B.pdf"),width = 4, height = 4)
print(a)
d <- dev.off()
a