pkgs <- c("ggthemes", "jhtools", "glue", "ggsci", "patchwork", "tidyverse",
"circlize", "ComplexHeatmap", "SummarizedExperiment", "jhuanglabRNAseq",
"viridis","ggrepel", "tidygraph","ggraph")
for (pkg in pkgs){
suppressPackageStartupMessages(library(pkg, character.only = T))
}
out_dir <- "./results/figure2" %>% checkdir
in_dir <- "./data" %>% checkdirfigure2
figure2A
group_twist2<- glue("{in_dir}/twist2_deg.rds") %>% read_rds
darw_volcano <- function(df){
tmp <- df %>% as_tibble(rownames = "genes") %>%
dplyr::mutate(label = case_when(
logFC > 1 & adj.P.Val < 0.05 ~ "UP",
logFC < -1 & adj.P.Val < 0.05 ~ "DOWN",
T ~ "NOSIG"
),
`- log10(adj.P.Val)` = - log10(adj.P.Val)) %>%
arrange(desc(abs(logFC)))
tmp$`- log10(adj.P.Val)`[tmp$`- log10(adj.P.Val)` > 75] <- 75
labeled_frame <- tmp %>%
dplyr::filter(abs(logFC) > 5.5 | - log10(adj.P.Val) > 45)
tmp %>%
ggplot(aes(x = logFC, y = `- log10(adj.P.Val)`, color = label)) +
geom_point(size = 0.01) +
scale_color_manual(values = c("UP" = "#DC143C", "DOWN" = "#1E90FF"), guide = guide_legend(
keywidth = 0.1)) +
theme_few() +
geom_vline(xintercept = c(1,-1), linetype = "dashed", size = 0.2) +
geom_hline(yintercept = c(1.30103), linetype = "dashed", size = 0.2) +
scale_y_continuous(expand = expansion(mult = c(.06, 0.15))) +
scale_x_continuous(expand = expansion(mult = c(.06, 0.15))) +
ggrepel::geom_label_repel(data = labeled_frame, mapping = aes(label = genes),
color = "black", fill = NA,
size = 2,
label.padding = 0.1,
max.overlaps = 1000,
label.r = 0.11,
label.size = 0.1,
min.segment.length = 0.1,
max.iter = 65,
force = 1,
force_pull = 0.1,
segment.size = 0.1) +
theme(
axis.ticks = element_line(linewidth = 0.2),
axis.ticks.length = unit(0.1, "lines"),
axis.text = element_text(size = 7, colour = "black"),
axis.title = element_text(size = 7, colour = "black"),
legend.box.margin = margin(0,0,0,0),
legend.box.spacing = unit(1, "mm"),
legend.key.width = unit(1.5, "mm"),
legend.key.height = unit(1.5, "mm"),
legend.text = element_text(size = 7),
legend.title = element_text(size = 7),
legend.spacing = unit(0, 'cm'),
legend.margin = margin(0,0,0,0),
panel.border = element_rect(colour = "black")
)
}
pdf(glue("{out_dir}/figure2A.pdf"), width = 3, height = 3.5)
print(darw_volcano(group_twist2))
d <- dev.off()
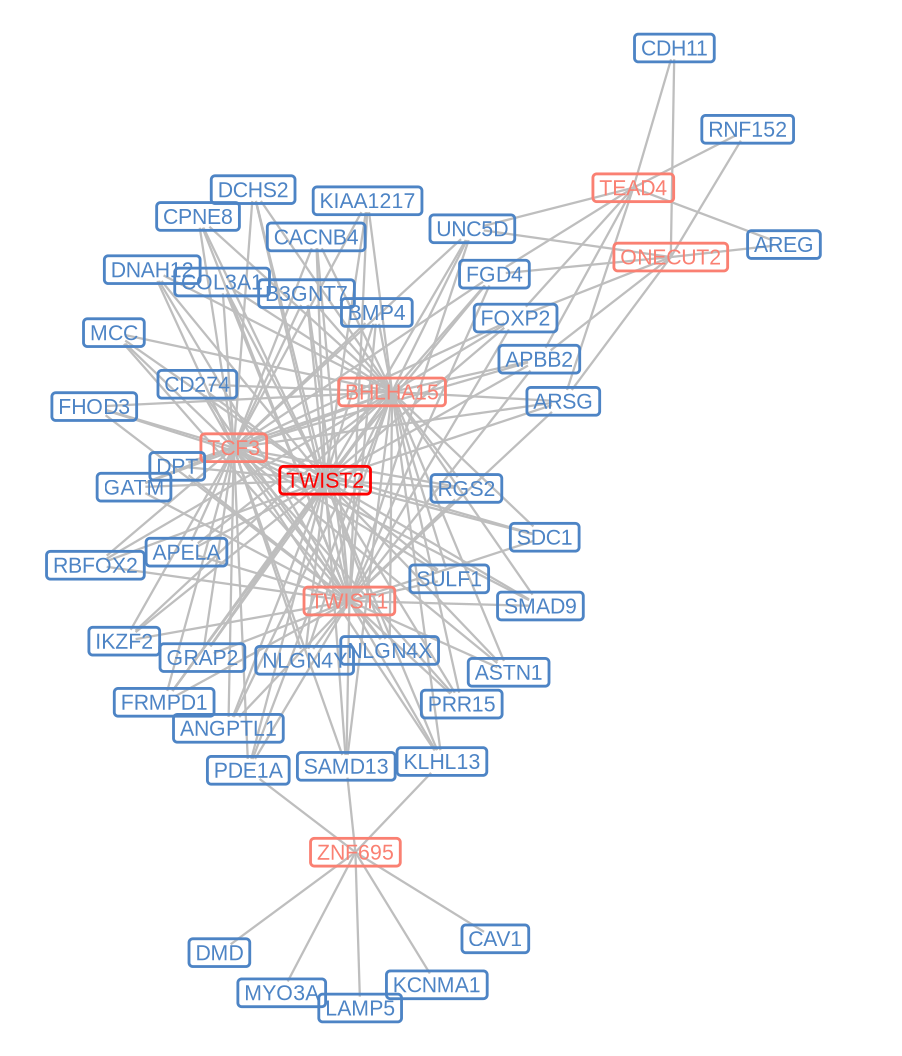
darw_volcano(group_twist2)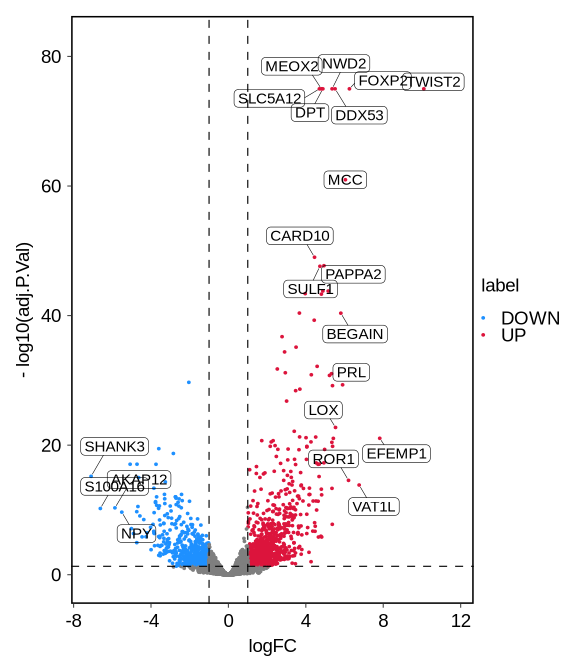
figure2B
sshal <- glue("{in_dir}/h.all.v2023.1.Hs.symbols.rds") %>% read_rds
a <- ggplot(sshal,aes(x = sub_types, y = gene_set %>% str_replace_all("_", " ") %>% str_wrap(width = 30),
fill = value)) +
geom_tile() +
scale_fill_gradient2(low = "blue", high = "red",
guide = guide_colourbar(label.hjust = 0.5)) +
theme_few() +
theme(
axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5,colour = "black"),
axis.text = element_text(size = 5,colour = "black"),
axis.ticks = element_line(linewidth = 0.2,colour = "black"),
axis.ticks.length = unit(0.05, "lines"),
axis.title = element_blank(),
legend.box.margin = margin(0,0,0,0),
legend.box.spacing = unit(1.5, "mm"),
legend.key.width = unit(2, "mm"),
legend.key.height = unit(2, "mm"),
legend.text = element_text(size = 5),
legend.title = element_text(size = 5),
legend.ticks.length = unit(0.1, "lines"),
panel.border = element_rect(colour = "black", linewidth = 0.3)
)
pdf(glue("{out_dir}/figure2B.pdf"), width = 4, height = 2)
print(a)
d <- dev.off()
a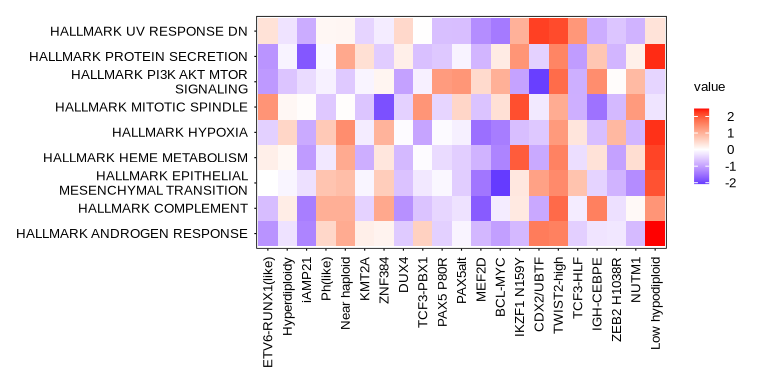
figure2C
subg <- read_rds(glue("{in_dir}/tf_net.rds"))
layoutx <- read_rds(glue("{in_dir}/tf_net_layout.rds"))
p0 <- ggraph(subg, layout = layoutx) +
geom_edge_link(end_cap = rectangle(3, 3, 'mm', 'mm'),
color = "grey", edge_width = 0.4) +
geom_node_label(aes(label = name, color = type),
repel = F, size = 2.8, fill = NA,
label.size = 0.5) +
scale_color_manual(values = c("A" = "#FA8072", "B" = "red", "C" = "#4D84C5")) +
theme_void() +
theme(legend.position = 'none')+
scale_x_continuous(limits = c(-2.8,2.4))
netoutdir <- checkdir("./paper_twist2_cdx2_g18/figure2/tf_enrich/net_figure")
pdf(glue("{out_dir}/figure2C.pdf"), width = 4.7, height = 5.5)
print(p0)
d <- dev.off()
p0