pkgs <- c("fs", "futile.logger", "configr", "stringr", "ggpubr", "ggthemes", "clusterProfiler",
"vroom", "jhtools", "glue", "openxlsx", "ggsci", "patchwork", "cowplot",
"tidyverse", "dplyr", "Seurat", "harmony", "ggpubr", "GSVA", "EnhancedVolcano")
suppressMessages(conflicted::conflict_scout())
for (pkg in pkgs){
suppressPackageStartupMessages(library(pkg, character.only = T))
}
res_dir <- "./results/figure6" %>% checkdir
dat_dir <- "./data" %>% checkdir
config_dir <- "./config" %>% checkdir
#colors config
config_fn <- glue::glue("{config_dir}/configs.yaml")
rctd_cols <- jhtools::show_me_the_colors(config_fn, "rctd_colors")
#read in coldata
sce_mac <- read_rds(glue::glue("{dat_dir}/sce_mac_DNp50_retsne.rds"))
spatial_ls <- read_rds(glue::glue("{dat_dir}/spatial_ls_rctd.rds"))figure6
figure6a
p <- DimPlot(sce_mac, reduction = "tsne", group.by = "cell_types2", pt.size = .001,
label = F, cols = c("DN-mp" = "#BC3C29FF", "Other-mp" = "#0072B5FF"), combine = F)
p <- p[[1]] +
guides(color = guide_legend(keywidth = unit(0, "cm"),
keyheight = unit(0, "cm")),
nrow = 2, byrow = T) +
theme(plot.title = element_blank(),
axis.text = element_blank(),
axis.title = element_text(size = 8),
axis.line = element_blank(),
axis.ticks = element_blank(),
plot.margin = margin(0,0,0,0),
legend.position = c(.05,.9),
legend.text = element_text(size = 6))
print(p)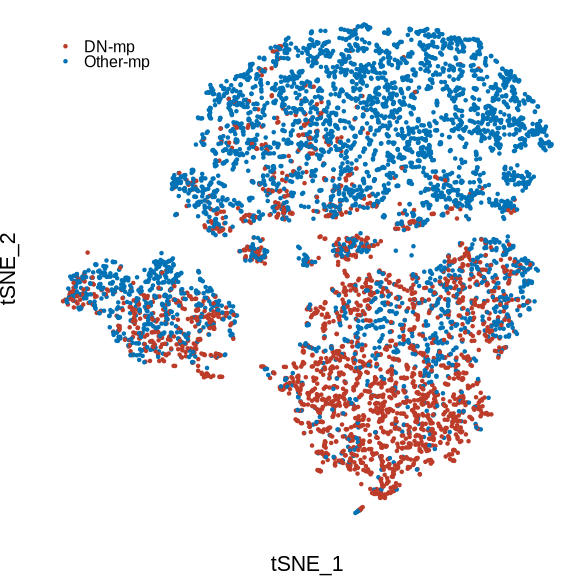
figure6b
p1 <- FeaturePlot(sce_mac, reduction = "tsne", features = "CD163",
order = T, pt.size = .001, combine = F)
p1 <- p1[[1]] +
guides(color = guide_colourbar(barwidth = unit(.1, "cm"),
barheight = unit(.6, "cm"),
title = "CD163",
title.position = "top")) +
theme(plot.title = element_blank(),
axis.text = element_blank(),
axis.title = element_text(size = 8),
axis.line = element_blank(),
axis.ticks = element_blank(),
panel.spacing = unit(0, "mm"),
panel.border = element_rect(fill = NA, colour = NA),
plot.background = element_rect(fill = NA, colour = NA),
plot.margin = margin(0,0,0,0),
legend.position = c(0,.8),
legend.margin = margin(0,0,0,0),
legend.title = element_text(size = 6),
legend.text = element_text(size = 6))
p2 <- FeaturePlot(sce_mac, reduction = "tsne", features = "HLA-DRB5",
order = T, pt.size = .001, combine = F)
p2 <- p2[[1]] +
guides(color = guide_colourbar(barwidth = unit(.1, "cm"),
barheight = unit(.6, "cm"),
title = "HLA-DRB5",
title.position = "top")) +
theme(plot.title = element_blank(),
axis.text = element_blank(),
axis.title = element_text(size = 8),
axis.line = element_blank(),
axis.ticks = element_blank(),
panel.spacing = unit(0, "mm"),
panel.border = element_rect(fill = NA, colour = NA),
plot.background = element_rect(fill = NA, colour = NA),
plot.margin = margin(0,0,0,0),
legend.position = c(0,.8),
legend.margin = margin(0,0,0,0),
legend.title = element_text(size = 6),
legend.text = element_text(size = 6))
p <- p1|p2
print(p)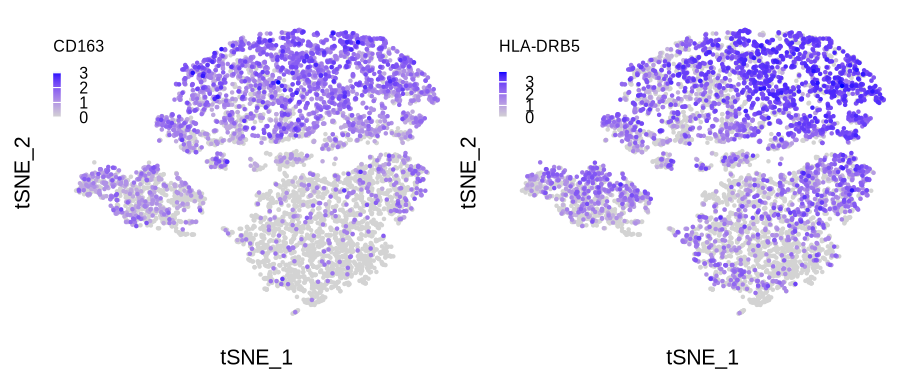
figure6c
p <- DotPlot(sce_mac, features = c('PTPRC', 'CD68', 'HLA-DRA', 'HLA-DRB1', 'HLA-DRB5', 'CD163'), scale = F,
dot.scale = 4.5) + RotatedAxis() + theme_bmbdc() +
scale_colour_gradientn(colours = c("grey", "#ffe5e5", "#ff7f7f", "#ff6666", "#ff0000"),
guide = guide_legend(keywidth = unit(100, "mm")),
values = scales::rescale(x = c(0, 1, 2, 3, 4), from = c(0, 4))) +
guides(size = guide_legend(title = "Percent Expressed", keywidth = unit(1, "cm"),
keyheight = unit(.2, "cm"), nrow = 2, byrow = T)) +
guides(color = guide_colorbar(title = "Expression", barwidth = unit(1, "cm"), barheight = unit(.2, "cm"))) +
theme(axis.text = element_text(size = 6),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
legend.key.size = unit(.2, 'cm'),
axis.text.x = element_text(angle = 30),
axis.text.y = element_text(angle = 50),
axis.line.x = element_line(linewidth = 0.4),
axis.line.y = element_line(linewidth = 0.4),
legend.margin = margin(0,0,0,0),
legend.title = element_text(size=6),
legend.text = element_text(size=6),
legend.position = "top")
print(p)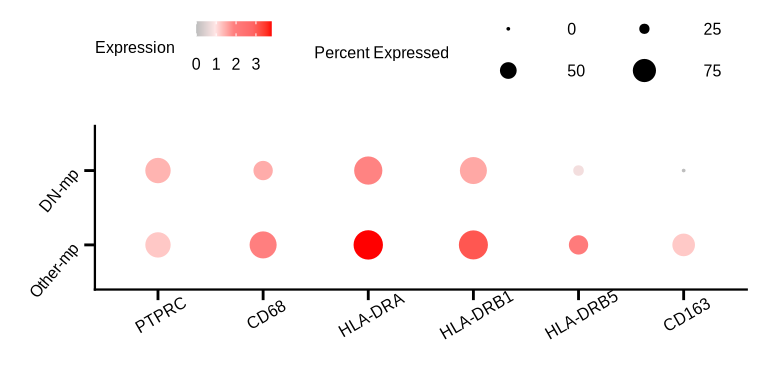
figure6d
de_df <- read_csv(glue::glue("{dat_dir}/pdac_DNvsOth_de_markers.csv")) %>% dplyr::filter(pct.1 > 0.1)
keyvals <- ifelse(
de_df$avg_log2FC < -0.5 & de_df$p_val_adj < 10e-10, "#0072B5FF",
ifelse(de_df$avg_log2FC > 0.5 & de_df$p_val_adj < 10e-10, "#BC3C29FF",
"#c7c1c1"))
keyvals[is.na(keyvals)] <- "#c7c1c1"
names(keyvals)[keyvals == "#0072B5FF"] <- "Other-mp"
names(keyvals)[keyvals == "#c7c1c1"] <- "NS"
names(keyvals)[keyvals == "#BC3C29FF"] <- "DN-mp"
p <- EnhancedVolcano(de_df,
lab = de_df$gene,
x = 'avg_log2FC',
y = 'p_val_adj',
pCutoff = 10e-10,
FCcutoff = 0.5,
pointSize = 1.5,
labSize = 1.5,
caption = "",
legendLabSize = 2.5,
legendIconSize = 1.5,
colAlpha = 0.8,
colCustom = keyvals) +
theme_bmbdc() +
theme(plot.title = element_blank(),
plot.subtitle = element_blank(),
text = element_text(size = 6),
axis.text = element_text(size = 6),
axis.title.x = element_text(size = 6),
axis.title.y = element_text(size = 6, vjust = -10),
axis.line.x = element_line(linewidth = 0.4),
axis.line.y = element_line(linewidth = 0.4),
legend.key.size = unit(.2, 'cm'),
legend.title = element_blank(),
legend.text = element_text(size=4),
legend.position = c(.75,.9)) +
guides(colour = guide_legend(ncol = 1))
ggsave(glue::glue("{res_dir}/fig6d_DNmac_pdac_deg_volcano.pdf"), p, width = 3, height = 4)
p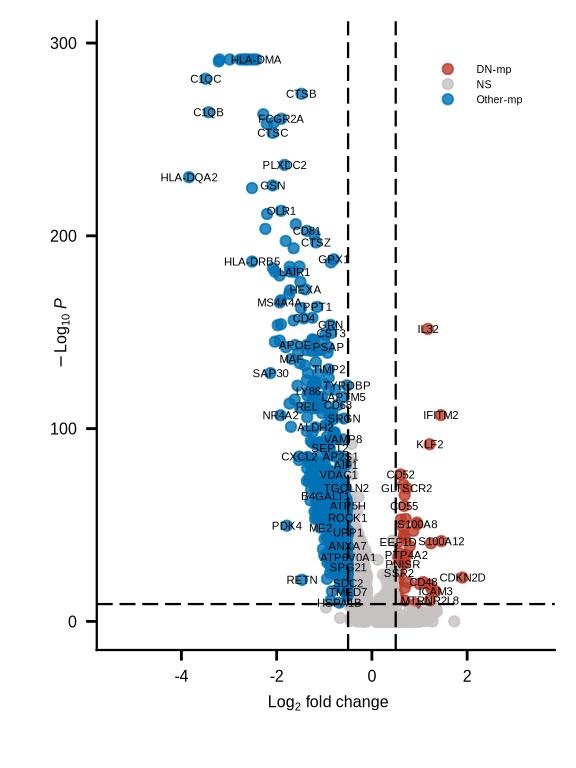
figure6e
gsea_df <- read_csv(glue::glue("{dat_dir}/goall_gsea_avg_mac.csv"))
p <- ggplot(data = gsea_df, aes(y = NES, x = gs_exact_source)) +
geom_col(fill = "#BC3C29FF") +
geom_text(aes(label = Description,
y = 0.1), size = 2, vjust = 0.5, hjust = 0) +
theme_bmbdc() +
theme(plot.title = element_blank(),
plot.subtitle = element_blank(),
text = element_text(size = 6),
axis.text = element_text(size = 6),
axis.title.y = element_blank(),
axis.title.x = element_text(size = 6),
axis.line.x = element_line(linewidth = 0.4),
axis.line.y = element_line(linewidth = 0.4),
legend.key.size = unit(.2, 'cm'),
legend.title = element_blank(),
legend.text = element_text(size=4),
legend.position = "top") +
coord_flip()
ggsave(glue::glue("{res_dir}/fig6e_DNmac_go_enrich.pdf"), p, height = 3, width = 4)
p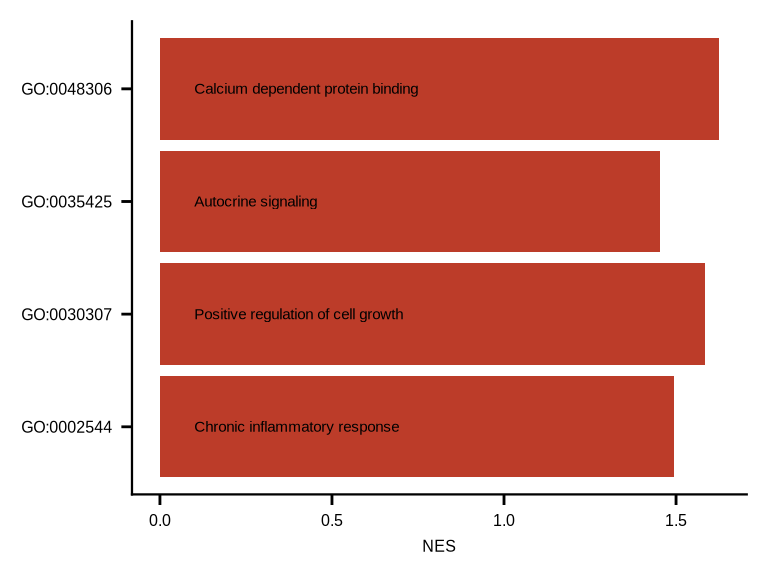
figure6f
p <- FeaturePlot(sce_mac, reduction = "tsne", features = "S100A9",
order = T, pt.size = .001, combine = F)
p <- p[[1]] +
guides(color = guide_colourbar(barwidth = unit(.1, "cm"),
barheight = unit(.6, "cm"),
title = "S100A9",
title.position = "top")) +
theme(plot.title = element_blank(),
axis.text = element_blank(),
axis.title = element_text(size = 8),
axis.line = element_blank(),
axis.ticks = element_blank(),
panel.spacing = unit(0, "mm"),
panel.border = element_rect(fill = NA, colour = NA),
plot.background = element_rect(fill = NA, colour = NA),
plot.margin = margin(0,0,0,0),
legend.position = c(0,.8),
legend.margin = margin(0,0,0,0),
legend.title = element_text(size = 6),
legend.text = element_text(size = 6))
print(p)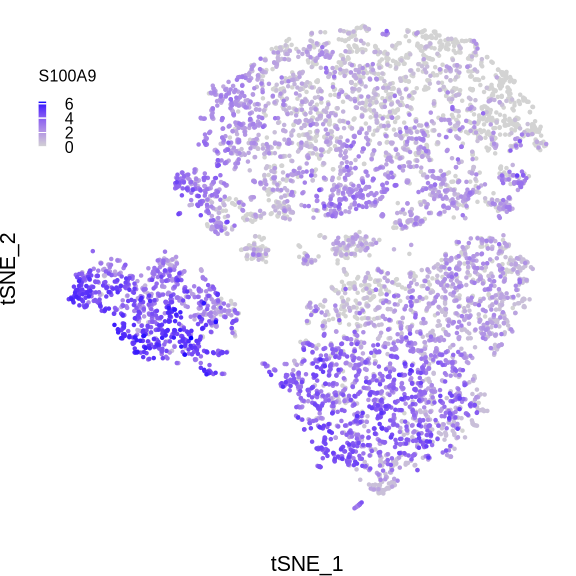
figure6g
#function
plot_deconv_pie <- function(rctd, deconv,
title = NULL,
my_pal = NULL,
size=0.35) {
my_table = rctd@spatialRNA@coords %>% as_tibble(rownames = "barcodes") %>%
dplyr::mutate(y1= y,
y = max(x) - x,
x = y1) %>% dplyr::select(-y1)
plot_val <- deconv %>% as_tibble(rownames = "barcodes")
my_table <- left_join(my_table, plot_val, by = "barcodes")
plot <- ggplot2::ggplot() +
scatterpie::geom_scatterpie(ggplot2::aes(x = x, y = y), data = my_table, pie_scale = size, col = NA,
cols = colnames(deconv)) +
coord_equal() + scale_fill_manual(values = my_pal) +
ggplot2::theme_classic() #+ coord_flip()
if(!is.null(title))
plot <- plot + ggplot2::ggtitle(title)
return(plot)
}
# images
p1 <- SpatialPlot(spatial_ls[["HTA12_25_5"]], alpha = c(0,0), combine = F)
p1 <- p1[[1]] +
coord_fixed() + theme_bmbdc() + theme(title = element_text(size = 6),
axis.text = element_text(size = 6, colour = "black"),
axis.title = element_blank(),
axis.ticks = element_line(colour = "black"),
plot.margin = margin(0.2,0.2,0.2,0.2),
legend.position = "none")
ggsave(glue::glue("{res_dir}/fig6g_image.pdf"), p1, width = 3.5, height = 3)
# deconv plot
rctd_ss <- read_rds(glue::glue("{dat_dir}/HTA12_25_5_RCTD.rds"))
df_decov_norm <- read_rds(glue::glue("{dat_dir}/df_decov_norm_HTA12_25_5.rds"))
p2 <- plot_deconv_pie(rctd = rctd_ss,
my_pal = rctd_cols,
deconv = df_decov_norm,
title = "HTA12_25_5") +
theme(title = element_text(size = 6),
axis.text = element_text(size = 6, colour = "black"),
axis.title = element_blank(),
axis.ticks = element_line(colour = "black"),
plot.margin = margin(0.2,0.2,0.2,0.2),
legend.margin = margin(0.1,0.1,0.1,0.1),
legend.text=element_text(size = 4),
legend.key.size = unit(.1, "cm"))
# pdf(glue::glue("{res_dir}/fig6g_spatial_deconv_pie.pdf"), width = 4, height = 3)
# print(p2)
# dev.off()
p1|p2 
figure6h
seu_sub <- read_rds(glue::glue("{dat_dir}/spatial_maconly_seu_obj.rds"))
p <- DotPlot(seu_sub, features = c('CD68', 'HLA-DRA', 'HLA-DRB1', 'HLA-DRB5', 'CD163'), scale = F,
dot.scale = 4.5, group.by = "DN_any") +
RotatedAxis() +
scale_colour_gradientn(colours = c("grey", "#ffe5e5", "#ff7f7f", "#ff6666", "#ff0000"),
values = scales::rescale(x = c(0, 0.5, 1, 1.5, 2), from = c(0, 2))) +
guides(size = guide_legend(title = "Percent Expressed", keywidth = unit(.2, "cm"),
keyheight = unit(.2, "cm"), nrow = 2, byrow = T)) +
guides(color = guide_colorbar(title = "Expression", barwidth = unit(1, "cm"), barheight = unit(.2, "cm"))) +
theme(plot.title = element_text(size = 6, face = "plain"),
axis.text = element_text(size = 6),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
legend.key.size = unit(.2, 'cm'),
axis.text.x = element_text(angle = 30),
axis.text.y = element_text(angle = 50),
axis.line.x = element_line(linewidth = 0.4),
axis.line.y = element_line(linewidth = 0.4),
legend.margin = margin(0,0,0,0),
legend.title = element_text(size=4),
legend.text = element_text(size=4),
legend.position = "top")
p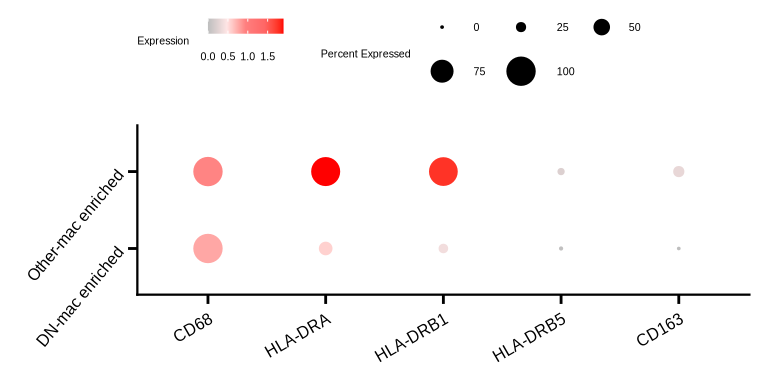
# pdf(file = glue::glue("{res_dir}/fig6h_seu_maconly_DNmacANY_markers_exp_noscale.pdf"),width = 4, height = 2)
# print(p)
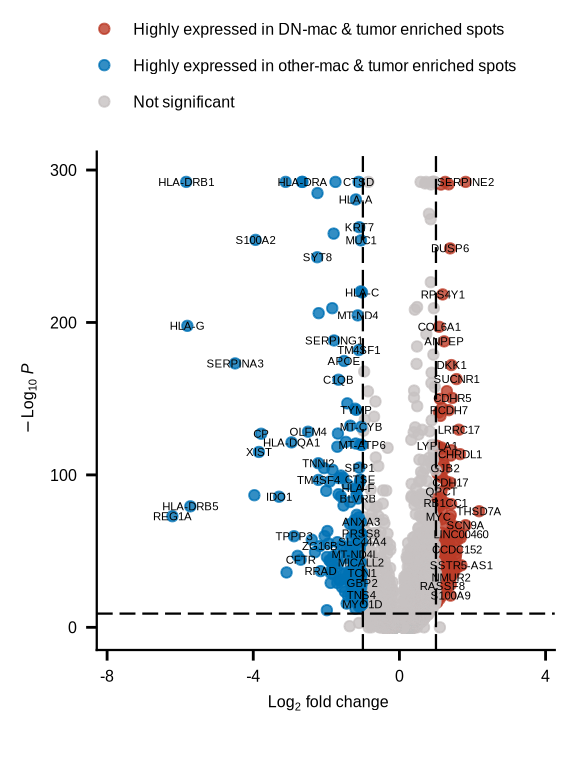
# dev.off()figure6i
de_markers <- read_csv(glue::glue("{dat_dir}/DN_any_sub_macDEGs.csv"))
keyvals <- ifelse(
de_markers$avg_log2FC < -1 & de_markers$p_val_adj < 10e-10, "#0072B5FF",
ifelse(de_markers$avg_log2FC > 1 & de_markers$p_val_adj < 10e-10, "#BC3C29FF",
"#c7c1c1"))
keyvals[is.na(keyvals)] <- "#c7c1c1"
names(keyvals)[keyvals == "#0072B5FF"] <- "Highly expressed in other-mac & tumor enriched spots"
names(keyvals)[keyvals == "#c7c1c1"] <- "Not significant"
names(keyvals)[keyvals == "#BC3C29FF"] <- "Highly expressed in DN-mac & tumor enriched spots"
p <- EnhancedVolcano(de_markers,
lab = de_markers$gene,
x = 'avg_log2FC',
y = 'p_val_adj',
pCutoff = 10e-10,
FCcutoff = 1,
pointSize = 1.5,
labSize = 1.5,
caption = "",
legendLabSize = 2.5,
legendIconSize = 1.5,
#legendPosition = "top",
colAlpha = 0.8,
colCustom = keyvals) +
theme_bmbdc() +
theme(plot.title = element_blank(),
plot.subtitle = element_blank(),
text = element_text(size = 6),
axis.text = element_text(size = 6),
axis.title.x = element_text(size = 6),
axis.title.y = element_text(size = 6),
axis.line.x = element_line(linewidth = 0.4),
axis.line.y = element_line(linewidth = 0.4),
legend.key.size = unit(.2, 'cm'),
legend.title = element_blank(),
legend.text = element_text(size=6),
legend.position = "top") +
guides(colour = guide_legend(nrow = 3))
ggsave(glue::glue("{res_dir}/fig6i_DNmac_visium_deg_volcano.pdf"), p, width = 3, height = 4)
p